Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN389-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN389-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 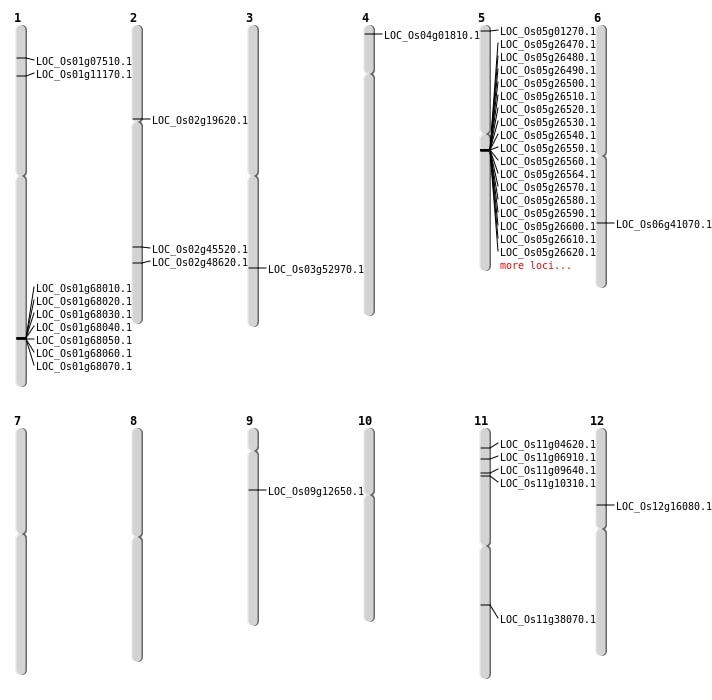
|
| Variant Information |
|---|
Single base substitutions: 44
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10361144 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 24757078 | G-A | HOMO | INTRON | |
| SBS | Chr1 | 25219404 | A-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 27172883 | C-A | HOMO | INTRON | |
| SBS | Chr1 | 5990995 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g11170 |
| SBS | Chr10 | 11892183 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 10974020 | A-G | HET | INTRON | |
| SBS | Chr11 | 17199702 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 18916858 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 1963973 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g04620 |
| SBS | Chr11 | 4963855 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 5832180 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 6159728 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 9781411 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 11733740 | C-T | HET | INTRON | |
| SBS | Chr12 | 14673412 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 16096555 | T-C | HET | INTRON | |
| SBS | Chr12 | 9181290 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os12g16080 |
| SBS | Chr12 | 9734314 | A-G | HET | UTR_3_PRIME | |
| SBS | Chr12 | 9736485 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 11458650 | C-T | HET | SPLICE_SITE_DONOR | LOC_Os02g19620 |
| SBS | Chr2 | 27699691 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g45520 |
| SBS | Chr2 | 29247359 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 3885992 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 22401245 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 28970912 | C-G | HET | INTRON | |
| SBS | Chr3 | 31342394 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 32249801 | G-C | HOMO | INTRON | |
| SBS | Chr4 | 13064750 | T-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 18655962 | G-A | HET | UTR_5_PRIME | |
| SBS | Chr4 | 31533820 | T-A | HET | INTRON | |
| SBS | Chr5 | 10137944 | C-G | HET | INTERGENIC | |
| SBS | Chr5 | 14264899 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 15001822 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 22476965 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 28043579 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 9890937 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 17186426 | G-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 30319125 | T-G | HOMO | INTERGENIC | |
| SBS | Chr7 | 10490348 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 25614988 | G-T | HET | INTRON | |
| SBS | Chr8 | 5862173 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 5862175 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 7241057 | G-T | HOMO | STOP_GAINED | LOC_Os09g12650 |
Deletions: 24
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 186912 | 186917 | 5 | LOC_Os05g01270 |
| Deletion | Chr4 | 522203 | 522205 | 2 | LOC_Os04g01810 |
| Deletion | Chr7 | 547547 | 547549 | 2 | |
| Deletion | Chr4 | 3502046 | 3502048 | 2 | |
| Deletion | Chr1 | 3587779 | 3587785 | 6 | LOC_Os01g07510 |
| Deletion | Chr11 | 5600949 | 5600956 | 7 | LOC_Os11g10310 |
| Deletion | Chr3 | 11859842 | 11859847 | 5 | |
| Deletion | Chr4 | 12115406 | 12115413 | 7 | |
| Deletion | Chr4 | 13428363 | 13428371 | 8 | |
| Deletion | Chr6 | 14204088 | 14204095 | 7 | |
| Deletion | Chr6 | 14231947 | 14231955 | 8 | |
| Deletion | Chr5 | 15387001 | 15937000 | 550000 | 86 |
| Deletion | Chr5 | 16119546 | 16119567 | 21 | |
| Deletion | Chr5 | 16345770 | 16345783 | 13 | |
| Deletion | Chr11 | 18487241 | 18487265 | 24 | |
| Deletion | Chr9 | 22615814 | 22615815 | 1 | |
| Deletion | Chr12 | 24431615 | 24431617 | 2 | |
| Deletion | Chr6 | 24531836 | 24531837 | 1 | LOC_Os06g41070 |
| Deletion | Chr12 | 25783823 | 25783833 | 10 | |
| Deletion | Chr2 | 29764063 | 29764066 | 3 | LOC_Os02g48620 |
| Deletion | Chr3 | 30386058 | 30386059 | 1 | LOC_Os03g52970 |
| Deletion | Chr3 | 31340650 | 31340666 | 16 | |
| Deletion | Chr1 | 39532503 | 39578748 | 46245 | 7 |
| Deletion | Chr1 | 41083517 | 41083528 | 11 |
Insertions: 5
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 3394885 | 3680217 | LOC_Os11g06910 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 5152576 | Chr2 | 29247130 | |
| Translocation | Chr11 | 5154876 | Chr2 | 29247094 | LOC_Os11g09640 |
| Translocation | Chr11 | 22578430 | Chr8 | 3761938 | LOC_Os11g38070 |
