Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN448-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN448-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 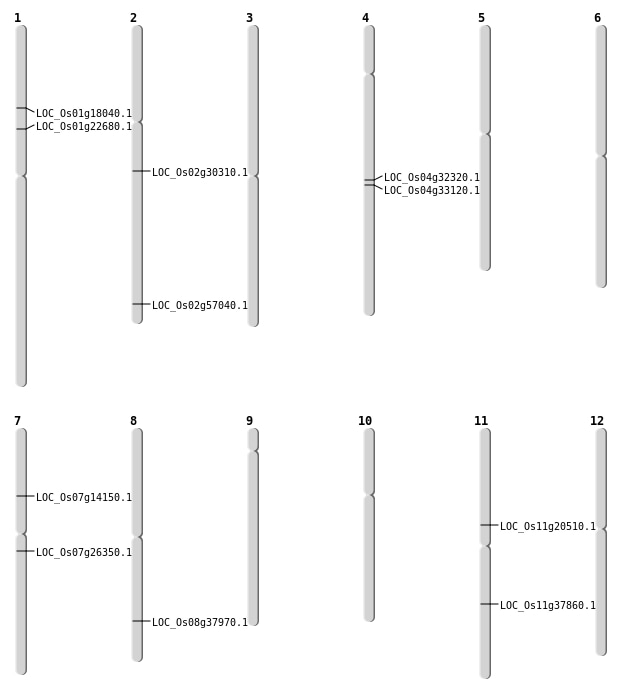
|
| Variant Information |
|---|
Single base substitutions: 35
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 18080199 | C-T | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr1 | 41818266 | A-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 41818270 | G-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 9587034 | C-T | HOMO | INTERGENIC | |
| SBS | Chr11 | 1096912 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 11933705 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 11970684 | A-T | HET | INTERGENIC | |
| SBS | Chr11 | 20699516 | A-T | HOMO | INTRON | |
| SBS | Chr11 | 21712681 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 23805056 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 26102732 | A-C | HET | INTRON | |
| SBS | Chr2 | 18031443 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 25648686 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 22934264 | G-A | HOMO | INTRON | |
| SBS | Chr3 | 2599867 | T-C | HET | INTRON | |
| SBS | Chr3 | 28661597 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 15080017 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 19376735 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g32320 |
| SBS | Chr4 | 20154698 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 23439474 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 28330131 | G-T | HOMO | INTERGENIC | |
| SBS | Chr5 | 19468513 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 11497493 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 13187539 | A-G | HET | INTRON | |
| SBS | Chr6 | 13856540 | T-G | HOMO | INTRON | |
| SBS | Chr6 | 29088630 | T-A | HET | INTRON | |
| SBS | Chr6 | 9363287 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 13034255 | C-T | HOMO | INTRON | |
| SBS | Chr7 | 15144770 | G-A | HET | STOP_GAINED | LOC_Os07g26350 |
| SBS | Chr7 | 19390031 | T-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 26203582 | G-A | HOMO | INTRON | |
| SBS | Chr8 | 13757954 | G-T | HET | INTRON | |
| SBS | Chr8 | 24052159 | G-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g37970 |
| SBS | Chr9 | 14561129 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 4856668 | A-G | HET | INTERGENIC |
Deletions: 18
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 435858 | 435859 | 1 | |
| Deletion | Chr12 | 1519697 | 1519698 | 1 | |
| Deletion | Chr4 | 1567057 | 1567062 | 5 | |
| Deletion | Chr6 | 1657679 | 1657687 | 8 | |
| Deletion | Chr11 | 4097385 | 4097386 | 1 | |
| Deletion | Chr7 | 8073404 | 8076332 | 2928 | LOC_Os07g14150 |
| Deletion | Chr9 | 11300763 | 11300775 | 12 | |
| Deletion | Chr2 | 15412838 | 15412839 | 1 | |
| Deletion | Chr4 | 16774309 | 16774329 | 20 | |
| Deletion | Chr2 | 18039674 | 18039675 | 1 | LOC_Os02g30310 |
| Deletion | Chr9 | 18221515 | 18221516 | 1 | |
| Deletion | Chr5 | 19655267 | 19655268 | 1 | |
| Deletion | Chr4 | 20053402 | 20053418 | 16 | LOC_Os04g33120 |
| Deletion | Chr5 | 20378428 | 20378432 | 4 | |
| Deletion | Chr11 | 20987461 | 20987462 | 1 | |
| Deletion | Chr8 | 22933499 | 22933523 | 24 | |
| Deletion | Chr3 | 30873413 | 30873416 | 3 | |
| Deletion | Chr1 | 33427139 | 33427140 | 1 |
Insertions: 7
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 9923879 | 11885986 | LOC_Os11g20510 |
| Inversion | Chr1 | 10096090 | 12751944 | 2 |
| Inversion | Chr11 | 22418926 | 22432951 | LOC_Os11g37860 |
| Inversion | Chr11 | 22418941 | 22433094 | LOC_Os11g37860 |
| Inversion | Chr2 | 34931316 | 35216202 | LOC_Os02g57040 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 4820787 | Chr9 | 18095067 | |
| Translocation | Chr7 | 5048173 | Chr3 | 19437692 | |
| Translocation | Chr3 | 10902804 | Chr1 | 34373526 | |
| Translocation | Chr7 | 21189219 | Chr4 | 13727661 | |
| Translocation | Chr12 | 27049993 | Chr6 | 6604775 |
