Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN552-S [Download] |
| Generation | M2-SOS |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN552-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 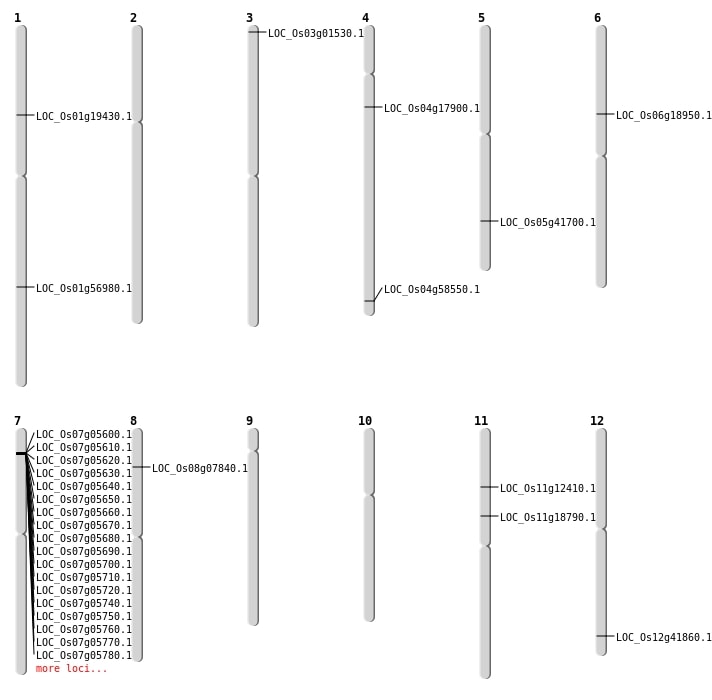
|
| Variant Information |
|---|
Single base substitutions: 34
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 15321121 | G-T | HOMO | INTRON | |
| SBS | Chr1 | 32913977 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g56980 |
| SBS | Chr10 | 11118370 | A-C | HET | INTERGENIC | |
| SBS | Chr10 | 12407968 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 10667584 | G-A | HOMO | STOP_GAINED | LOC_Os11g18790 |
| SBS | Chr12 | 15721677 | G-A | HET | INTRON | |
| SBS | Chr12 | 22106686 | G-A | HOMO | INTERGENIC | |
| SBS | Chr12 | 25923464 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g41860 |
| SBS | Chr12 | 25923465 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g41860 |
| SBS | Chr2 | 2086172 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 23896021 | C-A | HET | INTRON | |
| SBS | Chr2 | 5887166 | T-C | HOMO | INTERGENIC | |
| SBS | Chr2 | 5887169 | T-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 29934532 | T-A | HOMO | INTERGENIC | |
| SBS | Chr3 | 965832 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 31157438 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 6710422 | A-T | HET | INTRON | |
| SBS | Chr5 | 14449526 | G-A | HOMO | UTR_3_PRIME | |
| SBS | Chr5 | 24398049 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g41700 |
| SBS | Chr5 | 24398050 | C-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 8010857 | G-T | HOMO | INTERGENIC | |
| SBS | Chr6 | 10774758 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g18950 |
| SBS | Chr6 | 30352198 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 10906465 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g18430 |
| SBS | Chr7 | 10953197 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 26227125 | C-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 6960847 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 9385948 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 98242 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 27108401 | A-G | HOMO | INTRON | |
| SBS | Chr8 | 4399717 | T-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g07840 |
| SBS | Chr9 | 16235630 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 22313733 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 9236679 | G-C | HOMO | INTERGENIC |
Deletions: 25
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 341241 | 341242 | 1 | LOC_Os03g01530 |
| Deletion | Chr8 | 1095745 | 1095748 | 3 | |
| Deletion | Chr7 | 2253924 | 2253925 | 1 | |
| Deletion | Chr7 | 2629001 | 3292000 | 663000 | 101 |
| Deletion | Chr1 | 4649043 | 4649055 | 12 | |
| Deletion | Chr9 | 4679934 | 4679936 | 2 | |
| Deletion | Chr10 | 5900330 | 5900332 | 2 | |
| Deletion | Chr7 | 6961529 | 6961530 | 1 | |
| Deletion | Chr4 | 7358536 | 7358547 | 11 | |
| Deletion | Chr4 | 9670374 | 9670377 | 3 | |
| Deletion | Chr4 | 9844994 | 9844999 | 5 | LOC_Os04g17900 |
| Deletion | Chr1 | 10990866 | 10990884 | 18 | LOC_Os01g19430 |
| Deletion | Chr6 | 12307939 | 12307940 | 1 | |
| Deletion | Chr7 | 16172683 | 16172684 | 1 | |
| Deletion | Chr2 | 16579031 | 16579034 | 3 | |
| Deletion | Chr10 | 21071398 | 21071399 | 1 | |
| Deletion | Chr5 | 21271121 | 21271122 | 1 | |
| Deletion | Chr7 | 22322722 | 22322723 | 1 | |
| Deletion | Chr6 | 22967305 | 22967314 | 9 | |
| Deletion | Chr8 | 23146456 | 23146457 | 1 | |
| Deletion | Chr2 | 23510158 | 23510170 | 12 | |
| Deletion | Chr5 | 24514493 | 24514496 | 3 | |
| Deletion | Chr8 | 26662131 | 26662144 | 13 | |
| Deletion | Chr4 | 34810564 | 34810565 | 1 | LOC_Os04g58550 |
| Deletion | Chr3 | 35343984 | 35343990 | 6 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 12437994 | 12437994 | 1 | |
| Insertion | Chr11 | 17850788 | 17850788 | 1 | |
| Insertion | Chr5 | 1778871 | 1778873 | 3 | |
| Insertion | Chr7 | 11629690 | 11629703 | 14 | LOC_Os07g20135 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 6058722 | 6949544 | LOC_Os07g11010 |
| Inversion | Chr7 | 6058735 | 6700741 | LOC_Os07g11010 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 6927454 | Chr7 | 19968670 | LOC_Os11g12410 |
| Translocation | Chr4 | 11551033 | Chr2 | 17320497 | |
| Translocation | Chr8 | 16362992 | Chr4 | 18299491 |
