Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN601-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN601-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 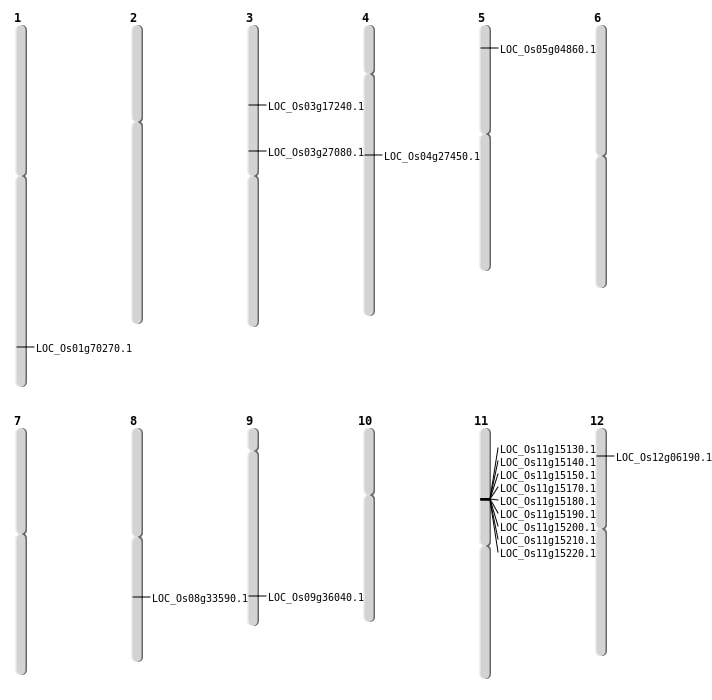
|
| Variant Information |
|---|
Single base substitutions: 29
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr10 | 11921231 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 12777268 | T-G | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr2 | 24912223 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 33668084 | G-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 10677023 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 19585872 | A-G | HET | INTRON | |
| SBS | Chr3 | 26963759 | G-C | HET | INTRON | |
| SBS | Chr3 | 33778499 | A-G | HET | INTRON | |
| SBS | Chr3 | 487107 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 9607356 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g17240 |
| SBS | Chr4 | 1908905 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 24319257 | T-A | HET | INTRON | |
| SBS | Chr4 | 25102166 | A-G | HET | UTR_5_PRIME | |
| SBS | Chr5 | 19510026 | T-C | HET | UTR_3_PRIME | |
| SBS | Chr5 | 5596276 | G-A | HOMO | INTERGENIC | |
| SBS | Chr5 | 7035034 | A-T | HOMO | INTRON | |
| SBS | Chr6 | 14838387 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 20406028 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 2721781 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 7522093 | C-A | HET | INTERGENIC | |
| SBS | Chr7 | 20783800 | G-A | HOMO | INTRON | |
| SBS | Chr7 | 28848212 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 8495693 | T-C | HOMO | INTERGENIC | |
| SBS | Chr8 | 11108762 | G-C | HOMO | INTERGENIC | |
| SBS | Chr8 | 12389009 | C-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 18376393 | C-A | HOMO | INTERGENIC | |
| SBS | Chr9 | 17325433 | T-G | HET | UTR_3_PRIME | |
| SBS | Chr9 | 19002150 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 20847694 | A-G | HET | INTERGENIC |
Deletions: 24
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 1660152 | 1660156 | 4 | |
| Deletion | Chr11 | 2483444 | 2483454 | 10 | |
| Deletion | Chr12 | 2935132 | 2935146 | 14 | LOC_Os12g06190 |
| Deletion | Chr1 | 5304298 | 5304370 | 72 | |
| Deletion | Chr6 | 5803341 | 5803344 | 3 | |
| Deletion | Chr11 | 8521001 | 8592000 | 71000 | 9 |
| Deletion | Chr6 | 12562345 | 12562355 | 10 | |
| Deletion | Chr10 | 13734465 | 13734467 | 2 | |
| Deletion | Chr12 | 14056948 | 14056949 | 1 | |
| Deletion | Chr7 | 15243441 | 15243450 | 9 | |
| Deletion | Chr9 | 15275024 | 15275027 | 3 | |
| Deletion | Chr3 | 15505659 | 15505660 | 1 | LOC_Os03g27080 |
| Deletion | Chr4 | 16233734 | 16233735 | 1 | LOC_Os04g27450 |
| Deletion | Chr2 | 18693542 | 18693543 | 1 | |
| Deletion | Chr9 | 20751997 | 20752145 | 148 | LOC_Os09g36040 |
| Deletion | Chr8 | 20974307 | 20974318 | 11 | LOC_Os08g33590 |
| Deletion | Chr5 | 22337920 | 22337928 | 8 | |
| Deletion | Chr7 | 22852358 | 22852368 | 10 | |
| Deletion | Chr1 | 23678682 | 23678979 | 297 | |
| Deletion | Chr7 | 25496048 | 25496049 | 1 | |
| Deletion | Chr7 | 26453306 | 26453315 | 9 | |
| Deletion | Chr4 | 28378448 | 28378451 | 3 | |
| Deletion | Chr2 | 32476293 | 32476294 | 1 | |
| Deletion | Chr1 | 40696310 | 40696314 | 4 | LOC_Os01g70270 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr2 | 17326875 | 17326876 | 2 | |
| Insertion | Chr3 | 905502 | 905502 | 1 | |
| Insertion | Chr5 | 2338785 | 2338785 | 1 | LOC_Os05g04860 |
| Insertion | Chr6 | 30067792 | 30067792 | 1 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 2725618 | 8617346 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 8941254 | Chr1 | 30237199 |
