Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN684-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN684-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 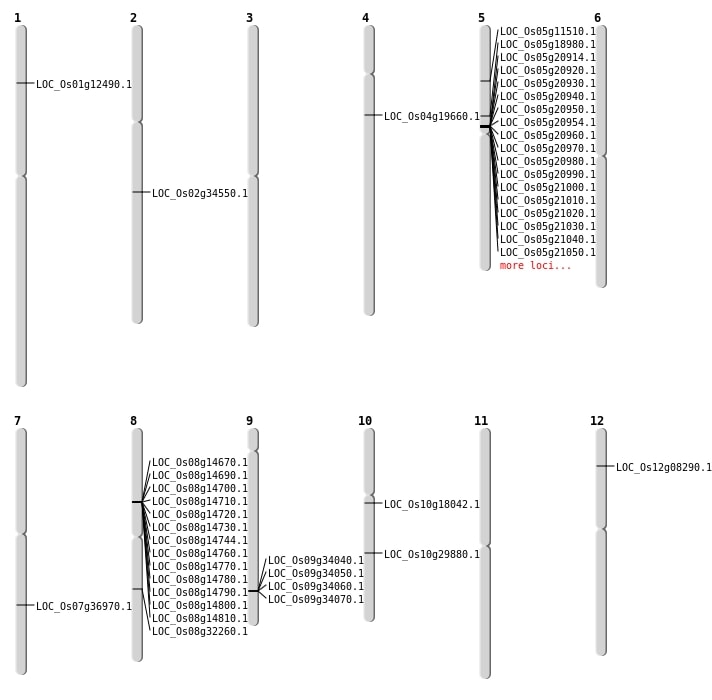
|
| Variant Information |
|---|
Single base substitutions: 34
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 27753968 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 31995305 | T-G | HOMO | INTRON | |
| SBS | Chr1 | 4372484 | A-G | HOMO | INTERGENIC | |
| SBS | Chr1 | 6858106 | C-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g12490 |
| SBS | Chr1 | 6858107 | T-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os01g12490 |
| SBS | Chr10 | 17280677 | T-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 19390956 | A-T | HET | INTRON | |
| SBS | Chr10 | 9145501 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g18042 |
| SBS | Chr11 | 23739833 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 8567408 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 4221108 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g08290 |
| SBS | Chr2 | 13178508 | A-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 16481609 | G-A | HOMO | INTRON | |
| SBS | Chr3 | 19236403 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 31709402 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 11015989 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g18980 |
| SBS | Chr5 | 14365635 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 16090513 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 17519333 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 9396070 | G-T | HET | INTRON | |
| SBS | Chr6 | 5893640 | T-C | HET | INTRON | |
| SBS | Chr7 | 10114786 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 10114787 | G-C | HOMO | INTERGENIC | |
| SBS | Chr7 | 10214767 | T-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 22132763 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g36970 |
| SBS | Chr7 | 23770635 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 25709391 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 24618450 | C-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 806254 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 12265310 | G-C | HET | INTRON | |
| SBS | Chr9 | 1471375 | G-T | HET | INTERGENIC | |
| SBS | Chr9 | 2241264 | G-T | HET | INTRON | |
| SBS | Chr9 | 2379634 | G-T | HET | INTERGENIC | |
| SBS | Chr9 | 2379635 | C-T | HET | INTERGENIC |
Deletions: 31
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 192297 | 192300 | 3 | |
| Deletion | Chr1 | 290262 | 290263 | 1 | |
| Deletion | Chr1 | 1782530 | 1782566 | 36 | |
| Deletion | Chr5 | 2983810 | 2983812 | 2 | |
| Deletion | Chr8 | 3249090 | 3249091 | 1 | |
| Deletion | Chr12 | 3493385 | 3493386 | 1 | |
| Deletion | Chr10 | 4747623 | 4747624 | 1 | |
| Deletion | Chr10 | 4747842 | 4747844 | 2 | |
| Deletion | Chr5 | 6515521 | 6515525 | 4 | LOC_Os05g11510 |
| Deletion | Chr8 | 8827001 | 8913000 | 86000 | 13 |
| Deletion | Chr8 | 9733423 | 9733424 | 1 | |
| Deletion | Chr4 | 10967440 | 10967444 | 4 | LOC_Os04g19660 |
| Deletion | Chr5 | 12103672 | 12103673 | 1 | |
| Deletion | Chr5 | 12283001 | 12417000 | 134000 | 21 |
| Deletion | Chr4 | 14560348 | 14560367 | 19 | |
| Deletion | Chr4 | 14882615 | 14882776 | 161 | |
| Deletion | Chr9 | 15328554 | 15328557 | 3 | |
| Deletion | Chr3 | 15498183 | 15498196 | 13 | |
| Deletion | Chr10 | 15524602 | 15524611 | 9 | LOC_Os10g29880 |
| Deletion | Chr3 | 15921528 | 15921529 | 1 | |
| Deletion | Chr12 | 17167341 | 17167342 | 1 | |
| Deletion | Chr1 | 17673962 | 17675372 | 1410 | |
| Deletion | Chr4 | 18982386 | 18982387 | 1 | |
| Deletion | Chr9 | 20089001 | 20108000 | 19000 | 4 |
| Deletion | Chr2 | 20713530 | 20713548 | 18 | LOC_Os02g34550 |
| Deletion | Chr12 | 21129620 | 21129621 | 1 | |
| Deletion | Chr1 | 26042230 | 26042235 | 5 | |
| Deletion | Chr4 | 27611250 | 27611268 | 18 | |
| Deletion | Chr3 | 30557064 | 30557067 | 3 | |
| Deletion | Chr1 | 31837854 | 31837866 | 12 | |
| Deletion | Chr2 | 34048888 | 34048895 | 7 |
Insertions: 5
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 12280851 | 12304550 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 2998740 | Chr1 | 37755113 | |
| Translocation | Chr8 | 19985564 | Chr1 | 17675367 | LOC_Os08g32260 |
