Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN689-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN689-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 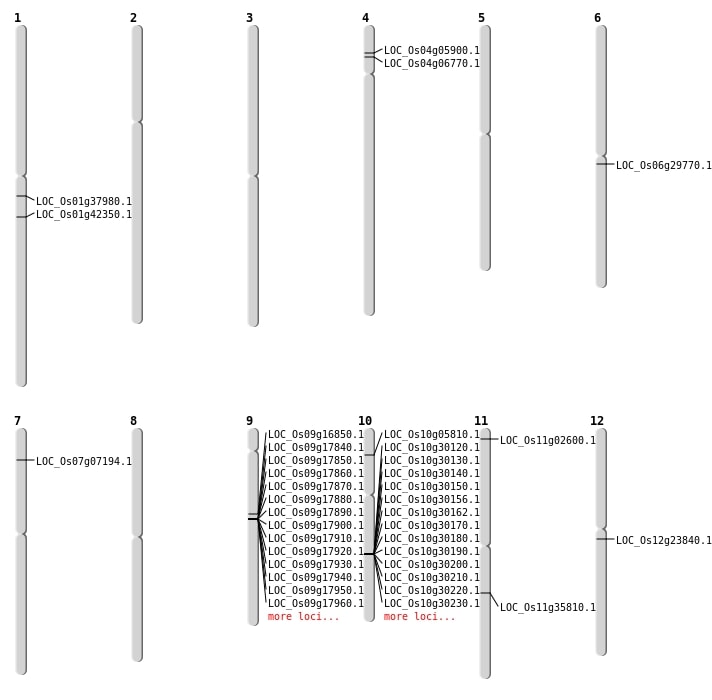
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 20069633 | C-T | HOMO | INTERGENIC | |
| SBS | Chr1 | 35271268 | G-C | HET | INTERGENIC | |
| SBS | Chr1 | 6375782 | A-G | HOMO | INTERGENIC | |
| SBS | Chr10 | 1629306 | G-A | HOMO | INTRON | |
| SBS | Chr10 | 21815530 | A-C | HOMO | INTERGENIC | |
| SBS | Chr10 | 2938924 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g05810 |
| SBS | Chr10 | 3023243 | C-A | HET | INTERGENIC | |
| SBS | Chr11 | 20814605 | A-C | HOMO | INTERGENIC | |
| SBS | Chr11 | 21018611 | T-C | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr11 | 21018612 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os11g35810 |
| SBS | Chr11 | 23819319 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 2959153 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 2959154 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 13556360 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os12g23840 |
| SBS | Chr2 | 10697770 | G-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 10697772 | C-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 13150437 | G-A | HOMO | INTERGENIC | |
| SBS | Chr4 | 19258207 | A-C | HOMO | INTERGENIC | |
| SBS | Chr4 | 26196729 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 27524873 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 3053521 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g05900 |
| SBS | Chr4 | 4177960 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 7510813 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 11607181 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 29516187 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 4985635 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 14247548 | C-T | HOMO | INTERGENIC | |
| SBS | Chr8 | 18419223 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 11341555 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os09g18500 |
| SBS | Chr9 | 11341557 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os09g18500 |
| SBS | Chr9 | 11341559 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os09g18500 |
| SBS | Chr9 | 13571830 | A-T | HOMO | INTRON | |
| SBS | Chr9 | 17972799 | A-T | HOMO | INTERGENIC |
Deletions: 27
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 1774804 | 1774805 | 1 | |
| Deletion | Chr11 | 2067619 | 2067627 | 8 | |
| Deletion | Chr11 | 3245359 | 3245363 | 4 | |
| Deletion | Chr4 | 3555832 | 3555833 | 1 | LOC_Os04g06770 |
| Deletion | Chr7 | 3566169 | 3566237 | 68 | LOC_Os07g07194 |
| Deletion | Chr8 | 3727296 | 3727304 | 8 | |
| Deletion | Chr6 | 9748522 | 9748531 | 9 | |
| Deletion | Chr11 | 10011566 | 10011573 | 7 | |
| Deletion | Chr9 | 10288001 | 10288029 | 28 | LOC_Os09g16850 |
| Deletion | Chr3 | 10655855 | 10655866 | 11 | |
| Deletion | Chr9 | 10912134 | 11184766 | 272632 | 35 |
| Deletion | Chr9 | 11184758 | 11387093 | 202335 | 32 |
| Deletion | Chr6 | 13357702 | 13357710 | 8 | |
| Deletion | Chr9 | 14441917 | 14441918 | 1 | |
| Deletion | Chr10 | 15648001 | 15717000 | 69000 | 15 |
| Deletion | Chr9 | 15862793 | 15862797 | 4 | LOC_Os09g26250 |
| Deletion | Chr6 | 17105895 | 17105937 | 42 | LOC_Os06g29770 |
| Deletion | Chr8 | 17687571 | 17687583 | 12 | |
| Deletion | Chr1 | 20067937 | 20067938 | 1 | |
| Deletion | Chr1 | 21278608 | 21278611 | 3 | LOC_Os01g37980 |
| Deletion | Chr10 | 21909489 | 21909507 | 18 | |
| Deletion | Chr11 | 22162746 | 22162752 | 6 | |
| Deletion | Chr7 | 23664273 | 23664274 | 1 | |
| Deletion | Chr1 | 24053292 | 24053300 | 8 | LOC_Os01g42350 |
| Deletion | Chr2 | 25470146 | 25470148 | 2 | |
| Deletion | Chr1 | 28833828 | 28833829 | 1 | |
| Deletion | Chr6 | 29485257 | 29485258 | 1 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 24574518 | 24574519 | 2 | |
| Insertion | Chr2 | 12550646 | 12550646 | 1 | |
| Insertion | Chr4 | 11939803 | 11939803 | 1 | |
| Insertion | Chr6 | 30941245 | 30941245 | 1 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 9374766 | 10912180 | |
| Inversion | Chr9 | 10267558 | 11387095 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 822541 | Chr4 | 16367930 | LOC_Os11g02600 |
