Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN722-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN722-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 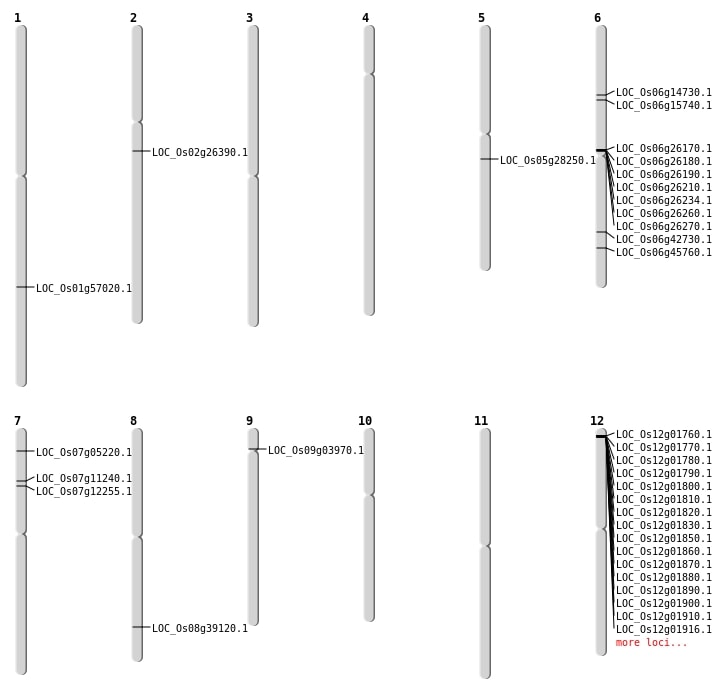
|
| Variant Information |
|---|
Single base substitutions: 32
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13552365 | C-T | HOMO | INTRON | |
| SBS | Chr1 | 28152347 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 35121130 | G-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 15348640 | T-A | HET | INTRON | |
| SBS | Chr10 | 15348641 | C-T | HET | INTRON | |
| SBS | Chr10 | 3739753 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 8605251 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr10 | 9469208 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 8451532 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 15062485 | C-G | HET | INTERGENIC | |
| SBS | Chr12 | 15276021 | G-C | HET | INTRON | |
| SBS | Chr12 | 21028253 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 25123812 | T-A | HOMO | INTERGENIC | |
| SBS | Chr2 | 29779336 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 16851999 | A-T | HOMO | INTRON | |
| SBS | Chr3 | 20136911 | C-T | HET | INTRON | |
| SBS | Chr5 | 11644980 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 11644981 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 3231761 | C-G | HOMO | INTRON | |
| SBS | Chr6 | 1453600 | C-A | HET | INTERGENIC | |
| SBS | Chr6 | 14825406 | C-A | HET | INTERGENIC | |
| SBS | Chr6 | 25701969 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 25701977 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os06g42730 |
| SBS | Chr7 | 2358535 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os07g05220 |
| SBS | Chr7 | 5718351 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 6716371 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 18487653 | C-G | HOMO | INTERGENIC | |
| SBS | Chr8 | 22043197 | C-T | HET | INTRON | |
| SBS | Chr8 | 24714310 | A-T | HOMO | STOP_GAINED | LOC_Os08g39120 |
| SBS | Chr9 | 12854597 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 13604391 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 3265524 | A-G | HOMO | INTERGENIC |
Deletions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 465001 | 938000 | 473000 | 90 |
| Deletion | Chr5 | 1805318 | 1805320 | 2 | |
| Deletion | Chr6 | 2038793 | 2038803 | 10 | |
| Deletion | Chr9 | 2041086 | 2041096 | 10 | LOC_Os09g03970 |
| Deletion | Chr6 | 2969316 | 2969322 | 6 | |
| Deletion | Chr2 | 4865190 | 4865199 | 9 | |
| Deletion | Chr12 | 5010647 | 5010667 | 20 | |
| Deletion | Chr7 | 5017148 | 5017149 | 1 | |
| Deletion | Chr7 | 6194110 | 6194124 | 14 | LOC_Os07g11240 |
| Deletion | Chr9 | 6670068 | 6670089 | 21 | |
| Deletion | Chr6 | 8299499 | 8299500 | 1 | LOC_Os06g14730 |
| Deletion | Chr3 | 9165141 | 9165142 | 1 | |
| Deletion | Chr10 | 12093467 | 12093469 | 2 | |
| Deletion | Chr6 | 15308001 | 15382000 | 74000 | 7 |
| Deletion | Chr2 | 15497000 | 15497015 | 15 | LOC_Os02g26390 |
| Deletion | Chr5 | 16541975 | 16541976 | 1 | LOC_Os05g28250 |
| Deletion | Chr1 | 18959454 | 18959462 | 8 | |
| Deletion | Chr4 | 20224267 | 20224268 | 1 | |
| Deletion | Chr2 | 20556227 | 20556234 | 7 | |
| Deletion | Chr11 | 27227127 | 27227128 | 1 | |
| Deletion | Chr12 | 27448285 | 27448300 | 15 | |
| Deletion | Chr6 | 27696430 | 27696433 | 3 | LOC_Os06g45760 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 7822042 | 7822043 | 2 | |
| Insertion | Chr11 | 19576681 | 19576683 | 3 | |
| Insertion | Chr3 | 24695999 | 24695999 | 1 | |
| Insertion | Chr6 | 20948219 | 20948219 | 1 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 6716611 | 6882638 | LOC_Os07g12255 |
Translocations: 8
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr7 | 226868 | Chr2 | 21806144 | |
| Translocation | Chr12 | 9725494 | Chr9 | 13612938 | |
| Translocation | Chr8 | 12894810 | Chr6 | 23394152 | |
| Translocation | Chr9 | 14377162 | Chr2 | 32007609 | |
| Translocation | Chr4 | 15070039 | Chr2 | 15004703 | |
| Translocation | Chr6 | 15382347 | Chr1 | 32942414 | 2 |
| Translocation | Chr6 | 15382466 | Chr1 | 32942160 | 2 |
| Translocation | Chr11 | 27773717 | Chr6 | 8926387 | LOC_Os06g15740 |
