Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN742-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN742-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 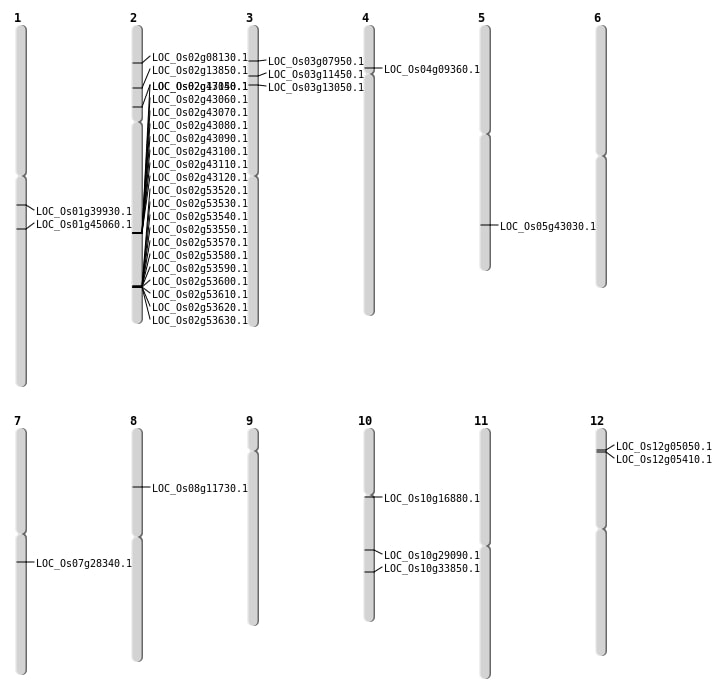
|
| Variant Information |
|---|
Single base substitutions: 39
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10404725 | A-G | HET | INTRON | |
| SBS | Chr1 | 18797630 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 22516214 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g39930 |
| SBS | Chr1 | 25581159 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os01g45060 |
| SBS | Chr1 | 6679067 | A-C | HET | INTERGENIC | |
| SBS | Chr10 | 6309330 | C-T | HOMO | SPLICE_SITE_REGION | |
| SBS | Chr11 | 10486420 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 11179822 | G-A | HOMO | INTERGENIC | |
| SBS | Chr11 | 13451842 | C-A | HET | INTRON | |
| SBS | Chr11 | 15413831 | G-T | HET | INTERGENIC | |
| SBS | Chr11 | 18310426 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 21142359 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 25070043 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 26392004 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 32356005 | T-A | HET | INTRON | |
| SBS | Chr2 | 32356006 | C-T | HET | INTRON | |
| SBS | Chr2 | 32456255 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 34696279 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 9816989 | G-A | HET | STOP_GAINED | LOC_Os02g17140 |
| SBS | Chr3 | 4066363 | C-T | HET | STOP_GAINED | LOC_Os03g07950 |
| SBS | Chr3 | 7062212 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g13050 |
| SBS | Chr4 | 12920825 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 21256962 | T-A | HET | INTERGENIC | |
| SBS | Chr4 | 2998393 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 7015641 | G-T | HET | INTRON | |
| SBS | Chr5 | 22285936 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 22658402 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 24898795 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 25569311 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 16545722 | C-G | HOMO | NON_SYNONYMOUS_CODING | LOC_Os07g28340 |
| SBS | Chr7 | 18072989 | G-C | HOMO | INTERGENIC | |
| SBS | Chr7 | 18226425 | G-A | HOMO | INTRON | |
| SBS | Chr7 | 5988991 | C-T | HOMO | INTRON | |
| SBS | Chr8 | 11454693 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 17486232 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 4943306 | G-A | HET | INTRON | |
| SBS | Chr8 | 6880636 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g11730 |
| SBS | Chr8 | 7581533 | A-T | HET | INTERGENIC | |
| SBS | Chr9 | 14992237 | A-G | HOMO | INTERGENIC |
Deletions: 26
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 1373508 | 1373509 | 1 | |
| Deletion | Chr6 | 2814585 | 2814587 | 2 | |
| Deletion | Chr5 | 3081148 | 3081149 | 1 | |
| Deletion | Chr9 | 4673546 | 4673548 | 2 | |
| Deletion | Chr4 | 4976275 | 4976299 | 24 | LOC_Os04g09360 |
| Deletion | Chr3 | 5900487 | 5900496 | 9 | LOC_Os03g11450 |
| Deletion | Chr4 | 7252893 | 7252897 | 4 | |
| Deletion | Chr2 | 7507724 | 7507725 | 1 | LOC_Os02g13850 |
| Deletion | Chr10 | 8413182 | 8413187 | 5 | LOC_Os10g16880 |
| Deletion | Chr2 | 9925716 | 9925721 | 5 | |
| Deletion | Chr11 | 12582328 | 12582332 | 4 | |
| Deletion | Chr11 | 14429968 | 14429971 | 3 | |
| Deletion | Chr12 | 17331898 | 17331971 | 73 | |
| Deletion | Chr10 | 17966063 | 17966075 | 12 | LOC_Os10g33850 |
| Deletion | Chr3 | 21356167 | 21356170 | 3 | |
| Deletion | Chr6 | 24643930 | 24643948 | 18 | |
| Deletion | Chr6 | 24647352 | 24647353 | 1 | |
| Deletion | Chr5 | 24988050 | 24988549 | 499 | LOC_Os05g43030 |
| Deletion | Chr6 | 25499839 | 25499842 | 3 | |
| Deletion | Chr2 | 25923001 | 25972000 | 49000 | 8 |
| Deletion | Chr8 | 26848524 | 26848532 | 8 | |
| Deletion | Chr2 | 27903695 | 27903698 | 3 | |
| Deletion | Chr3 | 32309854 | 32309855 | 1 | |
| Deletion | Chr2 | 32730001 | 32806000 | 76000 | 11 |
| Deletion | Chr1 | 40741357 | 40741360 | 3 | |
| Deletion | Chr1 | 42911892 | 42911895 | 3 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 23208034 | 23208036 | 3 | |
| Insertion | Chr5 | 20270791 | 20270791 | 1 | |
| Insertion | Chr7 | 4124573 | 4124574 | 2 | |
| Insertion | Chr9 | 10407045 | 10407046 | 2 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr12 | 2219329 | 2449993 | 2 |
| Inversion | Chr12 | 2223965 | 2450012 | LOC_Os12g05410 |
| Inversion | Chr4 | 4478005 | 20345712 | |
| Inversion | Chr10 | 18921378 | 19485347 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr2 | 4305744 | Chr1 | 10716869 | LOC_Os02g08130 |
| Translocation | Chr10 | 15151414 | Chr5 | 26317333 | LOC_Os10g29090 |
| Translocation | Chr5 | 24988168 | Chr2 | 4305748 | 2 |
| Translocation | Chr5 | 24988541 | Chr1 | 10717420 | LOC_Os05g43030 |
