Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN829-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN829-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 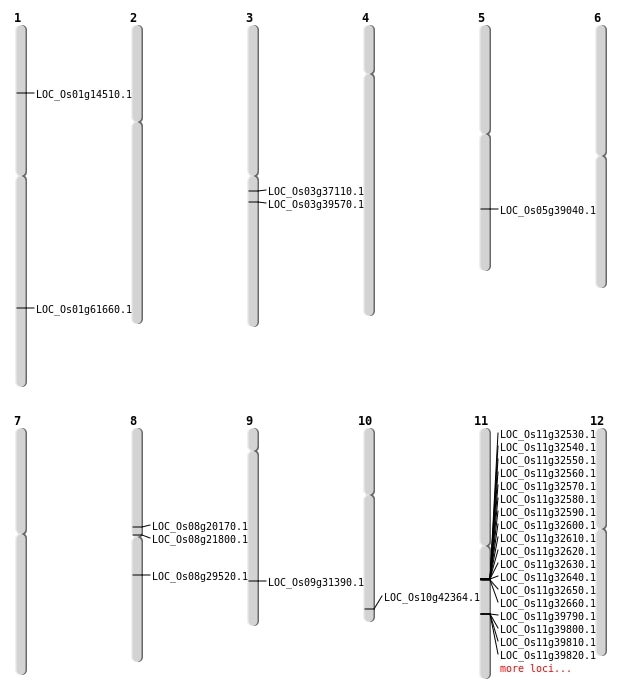
|
| Variant Information |
|---|
Single base substitutions: 25
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 17315952 | A-G | HET | INTRON | |
| SBS | Chr1 | 35677891 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g61660 |
| SBS | Chr1 | 36396299 | C-T | HET | INTRON | |
| SBS | Chr10 | 22805012 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g42364 |
| SBS | Chr11 | 20413851 | G-T | HET | INTRON | |
| SBS | Chr11 | 4244738 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 25034866 | C-G | HOMO | INTERGENIC | |
| SBS | Chr2 | 13078590 | C-T | HOMO | INTERGENIC | |
| SBS | Chr2 | 7537258 | A-C | HET | INTERGENIC | |
| SBS | Chr3 | 21971107 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g39570 |
| SBS | Chr3 | 21971108 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g39570 |
| SBS | Chr3 | 23797985 | A-T | HET | INTRON | |
| SBS | Chr3 | 4678172 | A-G | HOMO | INTERGENIC | |
| SBS | Chr3 | 4996160 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 2361885 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 6153902 | G-C | HET | INTERGENIC | |
| SBS | Chr4 | 8371328 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 22898705 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g39040 |
| SBS | Chr6 | 29862591 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 12405525 | A-T | HOMO | INTERGENIC | |
| SBS | Chr7 | 23742040 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 25473788 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 25718786 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 26098392 | T-A | HET | INTERGENIC | |
| SBS | Chr9 | 18875649 | T-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os09g31390 |
Deletions: 15
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 2215946 | 2215947 | 1 | |
| Deletion | Chr10 | 4542399 | 4542419 | 20 | |
| Deletion | Chr10 | 7448939 | 7448940 | 1 | |
| Deletion | Chr1 | 8127267 | 8127268 | 1 | LOC_Os01g14510 |
| Deletion | Chr8 | 9074518 | 9074525 | 7 | |
| Deletion | Chr8 | 12090378 | 12090386 | 8 | LOC_Os08g20170 |
| Deletion | Chr11 | 12649052 | 12649053 | 1 | |
| Deletion | Chr8 | 13048176 | 13048177 | 1 | LOC_Os08g21800 |
| Deletion | Chr11 | 15315918 | 15315923 | 5 | |
| Deletion | Chr8 | 18534940 | 18534943 | 3 | |
| Deletion | Chr11 | 19197001 | 19292000 | 95000 | 14 |
| Deletion | Chr11 | 23062687 | 23062688 | 1 | |
| Deletion | Chr11 | 23710001 | 23807000 | 97000 | 16 |
| Deletion | Chr3 | 30755479 | 30755488 | 9 | |
| Deletion | Chr3 | 33177300 | 33177306 | 6 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 22731681 | 22731681 | 1 | |
| Insertion | Chr11 | 26129615 | 26129616 | 2 | |
| Insertion | Chr5 | 6546094 | 6546094 | 1 | |
| Insertion | Chr5 | 979004 | 979004 | 1 |
Inversions: 8
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 2990366 | 3024958 | |
| Inversion | Chr8 | 17188900 | 18100772 | LOC_Os08g29520 |
| Inversion | Chr8 | 17188929 | 20751098 | |
| Inversion | Chr8 | 17189419 | 20751098 | |
| Inversion | Chr8 | 17189433 | 18100782 | LOC_Os08g29520 |
| Inversion | Chr11 | 19291871 | 21691230 | |
| Inversion | Chr11 | 19292071 | 21691225 | |
| Inversion | Chr3 | 20566862 | 20634239 | LOC_Os03g37110 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 13396680 | Chr3 | 5009447 |
