Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN908-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN908-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 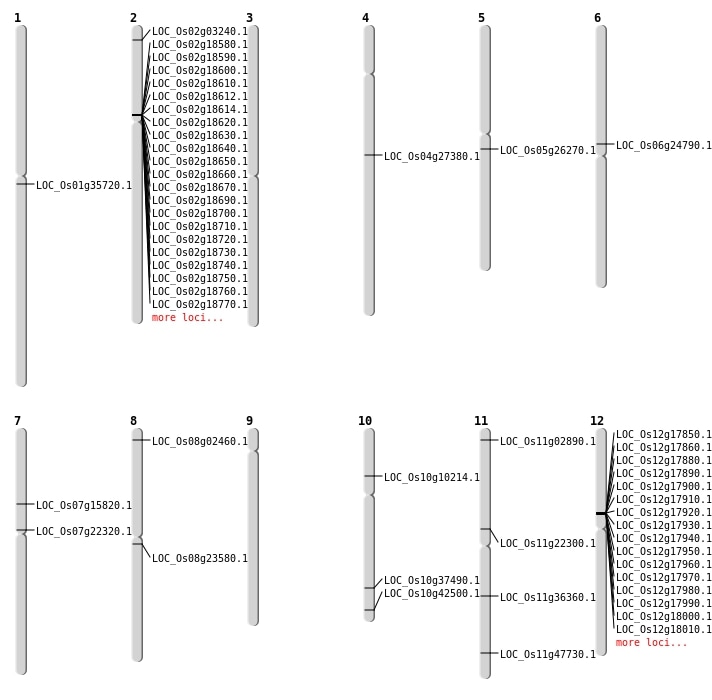
|
| Variant Information |
|---|
Single base substitutions: 39
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 19202459 | C-A | HET | INTERGENIC | |
| SBS | Chr1 | 22801554 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 18951419 | T-G | HET | INTERGENIC | |
| SBS | Chr10 | 5619500 | C-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os10g10214 |
| SBS | Chr11 | 12785857 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g22300 |
| SBS | Chr11 | 15882963 | G-C | HOMO | INTERGENIC | |
| SBS | Chr12 | 10034894 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 11691558 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 24242495 | C-T | HOMO | INTERGENIC | |
| SBS | Chr12 | 8827318 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 1297544 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g03240 |
| SBS | Chr2 | 2561566 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 3405171 | C-T | HET | INTRON | |
| SBS | Chr2 | 35471122 | G-A | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr2 | 5763180 | C-A | HET | INTRON | |
| SBS | Chr2 | 6007229 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 25933139 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 9863238 | T-C | HOMO | INTERGENIC | |
| SBS | Chr4 | 15193075 | A-C | HET | INTERGENIC | |
| SBS | Chr4 | 27408801 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 22894813 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 6372663 | T-C | HOMO | INTRON | |
| SBS | Chr5 | 8899875 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 15066808 | A-G | HET | INTRON | |
| SBS | Chr6 | 25510673 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 5122633 | T-A | HOMO | INTERGENIC | |
| SBS | Chr6 | 5751778 | G-A | HOMO | INTERGENIC | |
| SBS | Chr7 | 1065828 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 11126670 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 12513270 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g22320 |
| SBS | Chr7 | 4969099 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 8255422 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 9189037 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g15820 |
| SBS | Chr8 | 14271514 | G-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os08g23580 |
| SBS | Chr8 | 22283208 | G-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 469262 | C-T | HET | UTR_5_PRIME | |
| SBS | Chr9 | 12081052 | T-C | HET | INTRON | |
| SBS | Chr9 | 12339375 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 17685612 | A-G | HET | INTERGENIC |
Deletions: 19
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 4967143 | 4967158 | 15 | |
| Deletion | Chr10 | 5499340 | 5499349 | 9 | |
| Deletion | Chr12 | 8929046 | 8929050 | 4 | |
| Deletion | Chr11 | 9910985 | 9910992 | 7 | |
| Deletion | Chr12 | 10217001 | 10377000 | 160000 | 19 |
| Deletion | Chr2 | 10834001 | 10985000 | 151000 | 27 |
| Deletion | Chr5 | 12333641 | 12333651 | 10 | |
| Deletion | Chr3 | 12923666 | 12923671 | 5 | |
| Deletion | Chr11 | 14288030 | 14288034 | 4 | |
| Deletion | Chr2 | 18069617 | 18069665 | 48 | LOC_Os02g30330 |
| Deletion | Chr11 | 18925736 | 18925747 | 11 | |
| Deletion | Chr1 | 19190199 | 19190203 | 4 | |
| Deletion | Chr2 | 20087982 | 20087989 | 7 | |
| Deletion | Chr5 | 21094726 | 21094733 | 7 | |
| Deletion | Chr4 | 22597775 | 22597787 | 12 | |
| Deletion | Chr10 | 22923335 | 22923351 | 16 | LOC_Os10g42500 |
| Deletion | Chr5 | 27798790 | 27798791 | 1 | |
| Deletion | Chr4 | 29263934 | 29263940 | 6 | |
| Deletion | Chr2 | 34443828 | 34443840 | 12 | LOC_Os02g56280 |
Insertions: 11
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 23371282 | 23371283 | 2 | |
| Insertion | Chr1 | 40468997 | 40468999 | 3 | |
| Insertion | Chr11 | 28794547 | 28794554 | 8 | LOC_Os11g47730 |
| Insertion | Chr11 | 7087730 | 7087731 | 2 | |
| Insertion | Chr3 | 3312217 | 3312217 | 1 | |
| Insertion | Chr4 | 12689404 | 12689407 | 4 | |
| Insertion | Chr5 | 28034317 | 28034317 | 1 | |
| Insertion | Chr5 | 8964248 | 8964248 | 1 | |
| Insertion | Chr7 | 11319117 | 11319118 | 2 | |
| Insertion | Chr7 | 3577596 | 3577596 | 1 | |
| Insertion | Chr8 | 19998614 | 19998614 | 1 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 10985484 | 25468763 | 2 |
| Inversion | Chr2 | 10985651 | 25468758 | 2 |
| Inversion | Chr10 | 19468480 | 20071150 | LOC_Os10g37490 |
| Inversion | Chr1 | 24058572 | 24099211 |
Translocations: 8
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 986533 | Chr1 | 19784968 | 2 |
| Translocation | Chr11 | 7098218 | Chr4 | 13318315 | |
| Translocation | Chr9 | 9725009 | Chr5 | 15283670 | LOC_Os05g26270 |
| Translocation | Chr7 | 11905774 | Chr4 | 16178237 | LOC_Os04g27380 |
| Translocation | Chr11 | 14778496 | Chr8 | 985320 | LOC_Os08g02460 |
| Translocation | Chr12 | 16719731 | Chr6 | 13458685 | |
| Translocation | Chr3 | 19872591 | Chr2 | 13897067 | LOC_Os02g23970 |
| Translocation | Chr11 | 21424198 | Chr6 | 14556949 | 2 |
