Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | W463-2-1 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download W463-2-1 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 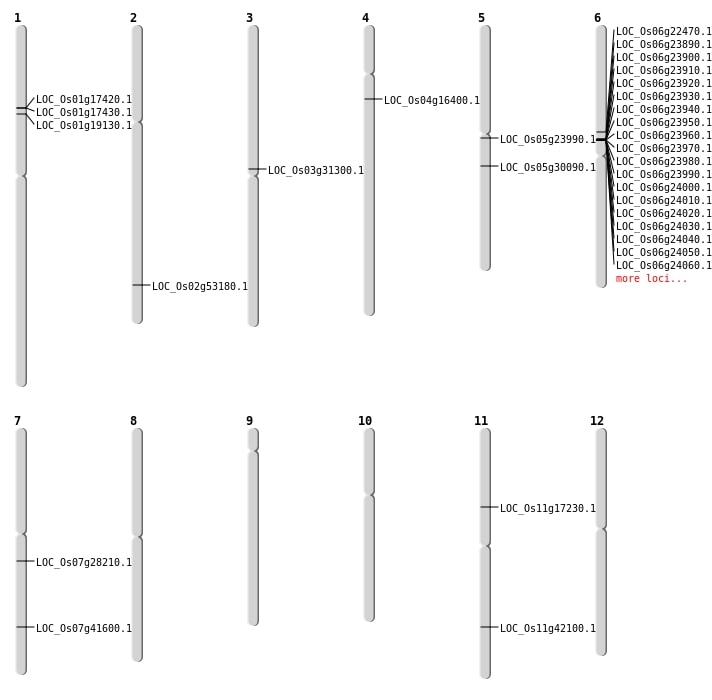
|
| Variant Information |
|---|
Single base substitutions: 41
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 1346163 | T-A | HET | UTR_5_PRIME | |
| SBS | Chr1 | 182278 | T-A | HOMO | INTERGENIC | |
| SBS | Chr1 | 19640305 | G-A | HOMO | INTRON | |
| SBS | Chr1 | 33937921 | A-G | HOMO | SYNONYMOUS_CODING | |
| SBS | Chr1 | 37251009 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 41165769 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 7600545 | T-A | HOMO | INTERGENIC | |
| SBS | Chr10 | 14666507 | G-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 14666508 | A-T | HOMO | INTERGENIC | |
| SBS | Chr10 | 20995899 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 671033 | G-A | HET | INTRON | |
| SBS | Chr11 | 8583489 | G-T | HET | INTERGENIC | |
| SBS | Chr11 | 9600208 | T-C | HOMO | NON_SYNONYMOUS_CODING | LOC_Os11g17230 |
| SBS | Chr12 | 12293327 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 13219902 | C-T | HET | INTRON | |
| SBS | Chr12 | 18828029 | A-C | HET | INTERGENIC | |
| SBS | Chr12 | 23480169 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 7845879 | T-G | HET | INTERGENIC | |
| SBS | Chr2 | 20119066 | A-C | HOMO | INTERGENIC | |
| SBS | Chr2 | 32629371 | C-T | HOMO | INTERGENIC | |
| SBS | Chr3 | 18111688 | T-A | HOMO | UTR_5_PRIME | |
| SBS | Chr3 | 23960800 | C-T | HET | INTRON | |
| SBS | Chr3 | 24401311 | C-G | HET | UTR_3_PRIME | |
| SBS | Chr3 | 26478365 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 31563434 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 36164513 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 8910473 | G-T | HOMO | NON_SYNONYMOUS_CODING | LOC_Os04g16400 |
| SBS | Chr5 | 13819575 | G-A | HOMO | NON_SYNONYMOUS_CODING | LOC_Os05g23990 |
| SBS | Chr5 | 17411746 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g30090 |
| SBS | Chr5 | 6432165 | T-C | HOMO | INTRON | |
| SBS | Chr5 | 9580956 | A-C | HOMO | INTERGENIC | |
| SBS | Chr6 | 12595622 | G-C | HOMO | INTERGENIC | |
| SBS | Chr7 | 1097667 | C-T | HOMO | INTRON | |
| SBS | Chr7 | 26305301 | T-G | HOMO | INTRON | |
| SBS | Chr8 | 16988827 | C-A | HOMO | UTR_5_PRIME | |
| SBS | Chr8 | 22747290 | T-G | HOMO | INTERGENIC | |
| SBS | Chr8 | 26636302 | C-A | HOMO | INTERGENIC | |
| SBS | Chr8 | 27882322 | C-G | HOMO | INTERGENIC | |
| SBS | Chr9 | 12211837 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 5384320 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 5540581 | T-C | HET | INTERGENIC |
Deletions: 24
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 2319385 | 2319402 | 17 | |
| Deletion | Chr8 | 3339205 | 3339206 | 1 | |
| Deletion | Chr5 | 5274532 | 5274535 | 3 | |
| Deletion | Chr5 | 6416410 | 6416421 | 11 | |
| Deletion | Chr4 | 7278748 | 7278751 | 3 | |
| Deletion | Chr8 | 7447379 | 7447380 | 1 | |
| Deletion | Chr1 | 10020019 | 10020046 | 27 | LOC_Os01g17420 |
| Deletion | Chr6 | 13061660 | 13061671 | 11 | LOC_Os06g22470 |
| Deletion | Chr6 | 13960248 | 13960250 | 2 | LOC_Os06g23890 |
| Deletion | Chr6 | 13962001 | 14255000 | 293000 | 44 |
| Deletion | Chr2 | 14693150 | 14693327 | 177 | |
| Deletion | Chr6 | 15409840 | 15409850 | 10 | |
| Deletion | Chr7 | 16478029 | 16478095 | 66 | LOC_Os07g28210 |
| Deletion | Chr11 | 17326621 | 17326628 | 7 | |
| Deletion | Chr3 | 18144507 | 18144620 | 113 | |
| Deletion | Chr1 | 18623503 | 18623538 | 35 | |
| Deletion | Chr1 | 19640284 | 19640296 | 12 | |
| Deletion | Chr9 | 21689475 | 21689491 | 16 | |
| Deletion | Chr1 | 21759427 | 21759428 | 1 | |
| Deletion | Chr3 | 23140818 | 23140819 | 1 | |
| Deletion | Chr5 | 23279078 | 23279085 | 7 | |
| Deletion | Chr2 | 25308797 | 25308799 | 2 | |
| Deletion | Chr11 | 25360006 | 25360016 | 10 | LOC_Os11g42100 |
| Deletion | Chr2 | 31061178 | 31061184 | 6 |
Insertions: 6
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 40840114 | 40840116 | 3 | |
| Insertion | Chr12 | 1185381 | 1185381 | 1 | |
| Insertion | Chr3 | 17832414 | 17832420 | 7 | LOC_Os03g31300 |
| Insertion | Chr6 | 29538984 | 29538984 | 1 | LOC_Os06g48810 |
| Insertion | Chr7 | 22523286 | 22523291 | 6 | |
| Insertion | Chr7 | 24934517 | 24934519 | 3 | LOC_Os07g41600 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 10024452 | 10807548 | 2 |
| Inversion | Chr1 | 10276223 | 10807534 | LOC_Os01g19130 |
| Inversion | Chr2 | 32559957 | 33310829 | LOC_Os02g53180 |
| Inversion | Chr2 | 32559969 | 33310850 | LOC_Os02g53180 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 25539245 | Chr1 | 40624405 |
